Abstract
Widespread use of gene therapy technologies is limited in part by the lack of small genetic switches with wide dynamic ranges that control transgene expression without the requirement of additional protein components1,2,3,4,5. In this study, we engineered a class of type III hammerhead ribozymes to develop RNA switches that are highly efficient at cis-cleaving mammalian mRNAs and showed that they can be tightly regulated by a steric-blocking antisense oligonucleotide. Our variant ribozymes enabled in vivo regulation of adeno-associated virus (AAV)-delivered transgenes, allowing dose-dependent and up to 223-fold regulation of protein expression over at least 43 weeks. To test the potential of these reversible on-switches in gene therapy for anemia of chronic kidney disease6, we demonstrated regulated expression of physiological levels of erythropoietin with a well-tolerated dose of the inducer oligonucleotide. These small, modular and efficient RNA switches may improve the safety and efficacy of gene therapies and broaden their use.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Our research resources, including methods, plasmids and protocols, are available upon reasonable request to qualified academic investigators for noncommercial research purposes. All reagents developed, including vector plasmids and detailed methods, will be made available upon written request. The corresponding author adheres to the NIH Grants Policy on Sharing of Unique Research Resources, including the ‘Sharing of Biomedical Research Resources: Principles and Guidelines for Recipients of NIH Grants and Contracts’.
References
Black, J. B., Perez-Pinera, P. & Gersbach, C. A. Mammalian synthetic biology: engineering biological systems. Annu. Rev. Biomed. Eng. 19, 249–277 (2017).
Alexander, H. K. et al. Selected technologies to control genes and their products for experimental and clinical purposes. Arch. Immunol. Ther. Exp. 55, 139–149 (2007).
Ye, X. et al. Regulated delivery of therapeutic proteins after in vivo somatic cell gene transfer. Science 283, 88–91 (1999).
Hjelm, B. E. et al. Mifepristone-inducible transgene expression in neural progenitor cells in vitro and in vivo. Gene Ther. 23, 424–437 (2016).
Favre, D. et al. Lack of an immune response against the tetracycline-dependent transactivator correlates with long-term doxycycline-regulated transgene expression in nonhuman primates after intramuscular injection of recombinant adeno-associated virus. J. Virol. 76, 11605–11611 (2002).
Hayat, A., Haria, D. & Salifu, M. O. Erythropoietin stimulating agents in the management of anemia of chronic kidney disease. Patient Prefer. Adherence 2, 195–200 (2008).
Auslander, S. & Fussenegger, M. Synthetic RNA-based switches for mammalian gene expression control. Curr. Opin. Biotechnol. 48, 54–60 (2017).
Tang, J. & Breaker, R. R. Rational design of allosteric ribozymes. Chem. Biol. 4, 453–459 (1997).
Yen, L. et al. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature 431, 471–476 (2004).
Win, M. N. & Smolke, C. D. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl Acad. Sci. USA 104, 14283–14288 (2007).
Ketzer, P., Haas, S. F., Engelhardt, S., Hartig, J. S. & Nettelbeck, D. M. Synthetic riboswitches for external regulation of genes transferred by replication-deficient and oncolytic adenoviruses. Nucleic Acids Res. 40, e167 (2012).
Beilstein, K., Wittmann, A., Grez, M. & Suess, B. Conditional control of mammalian gene expression by tetracycline-dependent hammerhead ribozymes. ACS Synth. Biol. 4, 526–534 (2015).
Zhong, G., Wang, H., Bailey, C. C., Gao, G. & Farzan, M. Rational design of aptazyme riboswitches for efficient control of gene expression in mammalian cells. eLife 5, e18858 (2016).
Mou, H. et al. Conditional regulation of gene expression by ligand-induced occlusion of a microRNA target sequence. Mol. Ther. 26, 1277–1286 (2018).
Wroblewska, L. et al. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 33, 839–841 (2015).
Wang, D., Tai, P. W. L. & Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 18, 358–378 (2019).
Scott, W. G., Horan, L. H. & Martick, M. The hammerhead ribozyme: structure, catalysis, and gene regulation. Prog. Mol. Biol. Transl. Sci. 120, 1–23 (2013).
Bellaousov, S., Reuter, J. S., Seetin, M. G. & Mathews, D. H. RNAstructure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 41, W471–W474 (2013).
Martick, M. & Scott, W. G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126, 309–320 (2006).
Bottaro, S. & Lindorff-Larsen, K. Mapping the universe of RNA tetraloop folds. Biophysical J. 113, 257–267 (2017).
Chi, Y. I. et al. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol. 6, e234 (2008).
Summerton, J. E. Morpholino, siRNA, and S-DNA compared: impact of structure and mechanism of action on off-target effects and sequence specificity. Curr. Top. Med Chem. 7, 651–660 (2007).
Stein, C. A. & Castanotto, D. FDA-approved oligonucleotide therapies in 2017. Mol. Ther. 25, 1069–1075 (2017).
Morcos, P. A., Li, Y. & Jiang, S. Vivo-morpholinos: a non-peptide transporter delivers morpholinos into a wide array of mouse tissues. Biotechniques 45, 613–614, 616, 618 (2008).
Summerton, J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta. 1489, 141–158 (1999).
Rivera, V. M. et al. Long-term regulated expression of growth hormone in mice after intramuscular gene transfer. Proc. Natl Acad. Sci. USA 96, 8657–8662 (1999).
Balazs, A. B. et al. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 481, 81–84 (2012).
Gardner, M. R. et al. AAV-expressed eCD4–Ig provides durable protection from multiple SHIV challenges. Nature 519, 87–91 (2015).
Huang, Y. Preclinical and clinical advances of GalNAc-decorated nucleic acid therapeutics. Mol. Ther. Nucleic Acids 6, 116–132 (2017).
Adamson, J. W. & Eschbach, J. W. Erythropoietin for end-stage renal disease. N. Engl. J. Med. 339, 625–627 (1998).
Lee, D. E., Son, W., Ha, B. J., Oh, M. S. & Yoo, O. J. The prolonged half-lives of new erythropoietin derivatives via peptide addition. Biochem. Biophys. Res. Commun. 339, 380–385 (2006).
Koulouridis, I., Alfayez, M., Trikalinos, T. A., Balk, E. M. & Jaber, B. L. Dose of erythropoiesis-stimulating agents and adverse outcomes in CKD: a metaregression analysis. Am. J. Kidney Dis. 61, 44–56 (2013).
Dunbar, C. E. et al. Gene therapy comes of age. Science 359, eaan4672 (2018).
Yen, L., Magnier, M., Weissleder, R., Stockwell, B. R. & Mulligan, R. C. Identification of inhibitors of ribozyme self-cleavage in mammalian cells via high-throughput screening of chemical libraries. RNA 12, 797–806 (2006).
Bohl, D., Salvetti, A., Moullier, P. & Heard, J. M. Control of erythropoietin delivery by doxycycline in mice after intramuscular injection of adeno-associated vector. Blood 92, 1512–1517 (1998).
Blum, S. et al. TARGT gene therapy platform for correction of anemia in end-stage renal disease. N. Engl. J. Med. 376, 189–191 (2017).
Macdougall, I. C. et al. Pharmacokinetics of novel erythropoiesis stimulating protein compared with epoetin alfa in dialysis patients. J. Am. Soc. Nephrol. 10, 2392–2395 (1999).
Chenuaud, P. et al. Autoimmune anemia in macaques following erythropoietin gene therapy. Blood 103, 3303–3304 (2004).
Gao, G. et al. Erythropoietin gene therapy leads to autoimmune anemia in macaques. Blood 103, 3300–3302 (2004).
Bunn, H. F. Drug-induced autoimmune red-cell aplasia. N. Engl. J. Med. 346, 522–523 (2002).
Acknowledgements
This work was supported by NIH R37 AI091476, NIH AI1129868 and NIH DP1 DA043912 (M.F.).
Author information
Authors and Affiliations
Contributions
G.Z. conceived this project and developed it with M.F. G.Z., H.W., Y.Y., and H.D. performed molecular cloning and cell culture experiments. G.Z., H.W., W.H., Y.L., H.M., Z.T., M.T. and T.O. performed animal experiments. G.Z. and M.F. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
G.Z. and M.F. are cofounders of Emmune Inc., a startup company that designs AAV-based treatments and prophylaxis for HIV-1.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
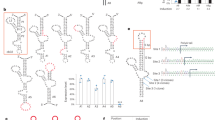
Supplementary Figure 1 Converting type I hammerhead ribozymes (HHR) to type III significantly improved ribozyme activity (from its original 18-fold to 130-fold).
(a) Sequences and secondary structures of three previously published type I hammerhead ribozyme variants, N79, N117, and N107 (Yen, L. et al. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature 431, 471-476, (2004).) and a stem I variant of N107 (N107-v1) are shown. Watson-Crick base-pairing interactions are indicated as orange lines. Other hydrogen-bonding interactions are represented, as in the main text, as thin black lines with annotations according to the following key: open circle next to open square represents a Watson-Crick/Hoogsteen interaction; open square next to open triangle, a Hoogsteen/sugar edge interaction; green line with T termini, nonadjacent base stacking; circle, a Wobble base pair. Thick solid or dashed orange lines indicate backbone continuity where the sequence has been separated for diagrammatic clarity. Thick blue lines indicate flexible linker sequences introduced to isolate the ribozyme and help stem-I folding. Arrows indicate a single A to G mutation that inactivates the ribozyme. Nucleotide differences among the variants are indicated in blue. Numbers in the parentheses are the fold inhibition activities of each variant when they are placed at 3′ UTR of a Gaussia luciferase (Gluc) reporter gene and tested in 293T cells using the reporter inhibition assay described in Fig. 1. (b) A potential explanation for the differences in reaction rates between a type I and type III HHR is diagramed. The enzyme strand of the HHR is indicated in orange, and the substrate strand is indicated in blue. A cross indicates the HHR cleavage site. The short substrate strand that dissociates after cleavage is referred as the leaving strand. The base-pairing interactions holding the long leaving strand of stem I and the tertiary interactions between this strand and loop II may slow the disassembly of the cleaved type I ribozyme and facilitate re-ligation of the cleaved substrate strand. In contrast, a cleaved type III HHR may disassemble more quickly with less re-ligation, due to its shorter leaving strand and the absence of tertiary interactions between its leaving strand and the remainder of the ribozyme. (c) Sequence and secondary structure of T3H1 ribozyme, a type III HHR converted in this study from the previously described HHR ribozyme, N107. Nucleotide differences from N107 ribozyme are indicated in blue. (d-e) N107-v1 and T3H1 were compared for their activities in a reporter inhibition assay, where a dual-reporter expression vector encoding a Gaussia luciferase (Gluc) and a Cypridina luciferase (Cluc), expressed from two independent promoters, was used. A catalytically active ribozyme variant was placed to the 3′ UTR (d) or 5’ UTR (e) of the Gluc gene, and compared with a catalytically inactive form of the ribozyme placed to the same sites. Reporter plasmids were transiently transfected into the indicated cell lines, and the ribozyme’s activity was calculated as fold inhibition in the Gluc expression relative to the inactive ribozyme control. A Cluc reporter was used as an internal control of Gluc expression. Consistent with the hypothesis represented in (b), type III ribozyme T3H1 outperformed type I ribozyme N107-v1 when placed at the 3′ UTR. Interestingly, no significant differences were observed when T3H1 and N107 were placed at the 5′ UTR, perhaps because cellular RNA helicases promote disassembly of both ribozymes. Thus ribozyme activity at the 5’UTR might indicate an initial cleavage rate independent of the rate of disassembly. Data shown are representative of two independent experiments with similar results, and data points represent mean ± s.d. of three cell cultures.
Supplementary Figure 2 Optimizing stem III sequences to balance cleavage and disassembly rates further improved ribozyme activity (from 130- to 300-fold).
(a) A diagram representing a proposed difference between enzymatic reactions of type III HHRs with different stem-III annealing energies. A more stable stem III facilitates cleavage but slows disassembly, whereas a less stable stem III works in the opposite direction. Thus an optimal stem III sequence that balances these processes affords the maximum inhibition of gene expression. The enzyme strand of the ribozyme is indicated in orange, and the substrate strand is indicated in blue. A cross indicates the cleavage site. (b-c) Stem-III sequences (b) and fold inhibition activities (c) of a panel of T3H1 variants, which differ from T3H1 only at its stem III region. Consistent with the hypothesis diagrammed in (a), ribozyme activity at the 3′ UTR increases as the stem III shortens from 8-bp to 5-bp, indicating that accelerated disassembly overcomes any loss of intrinsic catalytic activity. (d) The ribozyme variants in (b) were placed at the 5′ UTR, where the disassembly of a cleaved ribozyme may largely depend on the cellular RNA helicase activity. No significant change in ribozyme activity was observed when stem III was shortened from 8-bp to 6-bp. However, ribozyme cleavage activity decreased dramatically when the stem III was shortened from 6-bp to 4-bp, consistent with a loss of intrinsic activity with a less stable stem III. (e) Stem-III sequences of a panel of ribozyme variants, modified from those in (b) to have maximal numbers of potential inter-strand purine-base stackings in stem III. (f) The ribozyme variants in (e) were placed at the 3′ UTR and compared with the ribozymes in (b) for their fold inhibition activities in 293T cells. Note that a 6-bp stem III is optimal for both ribozyme panels, and that the 6-bp variant T3H16 exhibited the highest fold inhibition. (g-h) Stem-III sequences (g) and fold inhibition activities in 293T cells (h) of a panel of T3H16 variants which only differ in the number of potential inter-strand purine-base stackings. Ribozymes T3H16 and T3H29 showed the highest fold inhibition activity, ~300-fold. (i) The fold inhibition activities of the stem-III variants were plotted against their calculated stem-III annealing energies. Ribozyme activities show a negative skewed distribution, and both T3H16 and T3H29 have annealing energies of -9.15±0.05 kcal/mol. Data shown are representative of three independent experiments with similar results, and data points represent mean ± s.d. of three cell cultures.
Supplementary Figure 3 Stabilizing tertiary interactions between stem I and loop II further improved ribozyme activity (from 300-fold to 1200-fold).
(a) The secondary structure of a crystalized Schistosoma mansoni HHR and the tertiary structure of a loop II-interacting bulge in its stem I are shown (PDB: 3ZD5). The extensive tertiary interactions between this bulge and loop II are essential for the function of an HHR in mammalian cells (Khvorova, A., Lescoute, A., Westhof, E. & Jayasena, S. D. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol 10, 708-712, (2003); Martick, M. & Scott, W. G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126, 309-320, (2006)). Based on the crystal structure, another smaller bulge above the loop II-interacting bulge may destabilize these tertiary interactions We therefore sought to stabilize this loop II-interacting bulge through stem-I changes. (b) Stem-I sequences of a panel of T3H16 variants, which differ from T3H16 only in stem I. Loop II-interacting bulges are highlighted in orange. Numbers in the parentheses indicate fold-inhibition activities of the corresponding ribozymes in 293T cells. (c-d) Sequences (c) and fold inhibition activities in 293T cells (d) of a panel of ribozyme variants carrying nucleotide mutations (black) at the bulge I/loop II region. T3H54 and T3H56 are T3H44 variants bearing single-nucleotide changes designed to impair bulge I/loop II tertiary interactions, and serve here as controls. Similarly, T3H57 is a variant of T3H41 with a single, destabilizing nucleotide change. (e) The stem I sequences of a panel of T3H44 variants differing from T3H44 only in the loop regions indicated in black. Numbers in the parentheses indicate fold inhibition activities of the corresponding ribozymes in 293T cells. (f) Fold-inhibition activities in 293T cells of the indicated hammerhead ribozyme variants at the 5′ UTR or 3′ UTR of the Gluc reporter gene. (g-h) Sequences (g) and fold-inhibition activities in 293T cells (h) of N107, N117, three N107-derived type-III hammerhead ribozyme variants (T3H16, T3H36, T3H37), and three N117-derived type-III hammerhead ribozyme variants (T3H16×117, T3H36×117, T3H37×117). (i-j) Sequences (i) and fold inhibition activities in 293T cells (j) of four type-III hammerhead ribozyme variants. sTRSV-H1 is a natural type-III hammerhead ribozyme from satellite tobacco ringspot viral genome (Chi, Y. I. et al. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol 6, e234, (2008)). sTRSV-H2 is a ribozyme variant identical to sTRSV-H1 except that the stem-III sequence is replaced with the sequence from T3H16. Calculated stem-III annealing energy values for sTRSV-H1 and sTRSV-H2 are shown in parentheses. Data shown are representative of two or three independent experiments with similar results, and data points in panels d, f, h, and j represent mean ± s.d. of three (panel j) or four (panels d, f, and h) cell cultures.
Supplementary Figure 4 Cell-type dependent activities of ribozyme variants and the v-M8 morpholino.
(a) Fold-inhibition of reporter expression with the indicated ribozyme variants in five different cell lines. Note that the rank order of ribozyme regulation remains consistent across cell lines. Data points represent mean ± s.d. of eight cell cultures from two independent experiments. (b) T3H38 ribozyme and v-M8 morpholino were tested in the five indicated cell lines for induction of Gluc expression. Gluc expression levels were normalized against the expression from a control reporter construct that carries no ribozyme element. v-M8 morpholino-mediated fold induction of active T3H38-regulated Gluc expression is shown for each cell line. Data points represent mean ± s.e.m of two (A549) or three (293T, Huh7, HeLa, MEF) independent experiments, each with three cell cultures.
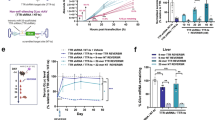
Supplementary Figure 5 In vivo regulation of adeno-associated virus (AAV)-delivered reporter expression in mouse muscle.
(a) Luciferase expression in the same mice shown in panel b of Fig. 3 was monitored over a 5-week period. Data points represent mean ± s.d. of three mice per group. (b) Bioluminescence images of mice used to generate the data of Fig. 3d. These experiments were independently repeated three times with similar results. (c) Eight-week old female BALB/c mice received bilateral intramuscular (i.m.) injections of T3H38-regulated AAV-Fluc at 5×109 GC/leg. Mice then received daily intravenous (i.v.) injections of PBS or 12.5 mg/kg v-M8 morpholino from day 7 to day 10 post AAV injection. In vivo bioluminescent imaging was performed at the indicated time points using the Xenogen IVIS In-Vivo Imager. (d) In vivo bioluminescent quantification data for the mice images in panel c. Data points represent mean ± s.d. of five mice per group.
Supplementary Figure 6 Off-target activity of the v-M8 morpholino.
(a) Sequences of v-M8 morpholino and its complement, v-M8′. (b) BLAST analysis was performed on the human transcriptome to identify transcript sequences similar to the v-M8′. Identifiers and sequence information of 11 potential off-target transcripts that are PCR-detectable in 293T cellular RNAs are listed. (c) RT-qPCR analysis for these 11 potential off-target transcripts using total RNA samples from 293T cells receiving no morpholino treatment, or treated with 10 µM of a control morpholino (v-M3R) or the v-M8 morpholino. Relative quantification of each potential off-target transcript was performed using GAPDH as an internal control. The abundance of each transcript was normalized to the abundance of the same transcript detected in the “No morpholino” control samples. v-M8 morpholino treatment significantly increased abundance of the SEMA6A-AS2, GRSF1 transcript variant X1, SMIM10L1, and IKZF2 transcript variant X23 (two-sample Student’s t-test, two-sided, P = 0.0001, 0.036, 1.38×10-6, and 0.018, repectively). Data points represent mean ± s.d. of four cell cultures.
Supplementary Figure 7 In vivo regulation of AAV-delivered reporter expression in mouse muscle and liver.
(a) Bioluminescence images of the two groups of mice used to generate data of Fig. 3e. Group-1 mice received i.m. injections of 0.5 mg/kg v-NC, 0.5 mg/kg v-M8, 0.5 mg/kg v-M8, and 2.5 mg/kg v-M8 at weeks 2, 7, 15, and 38 post AAV injection, respectively. Group-2 mice received i.m. injections of 0.5, 0.5, and 2.5 mg/kg v-M8 at weeks 2, 7, and 38 post AAV injection, respectively. In vivo bioluminescent images for the indicated time points are shown, with imaging photos from a same morpholino induction cycle shown in a row. (b) The same mice used in panel a were i.m. injected again with 2.5 mg/kg v-M8 morpholino at day 600 post AAV injection. In vivo bioluminescent imaging was performed at the indicated time points. (c) Six- to eight-week old BALB/c or C57BL/6J mice were injected with 1×1010 GC of catalytically active or inactive T3H38-regulated AAV-Fluc. I.m. injection of AAV1 particles was performed for muscle transduction, while i.v. injection of AAV8 particles was performed for liver transduction. In vivo bioluminescent imaging was performed at week 1 post AAV injection. Each bar indicates the mean and range of the luciferase signals of each mouse group. The numbers of mice used in each group are indicated next to each bar. (d-e) Eight-week old female BALB/c mice were i.v. injected with 1x1010 GC of AAV particles expressing firefly luciferase regulated by an active T3H38 ribozyme. Seventeen days later, mice were injected i.v. daily with PBS or 12.5 mg/kg of v-M8 morpholino for four days. Luciferase expression at the indicated time points was measured using the Lago X optical imaging system. Image data (d) and quantification of bioluminescence (e) are shown. Data points represent mean ± s.d. of four mice per group.
Supplementary Figure 8 In vivo regulation of AAV-delivered erythropoietin (Epo) expression.
(a) Individual data points for Fig. 4c, d were plotted. Each line represents Epo protein concentrations (red line) or hematocrit counts (blue line) measured using tail-vein blood samples collected from a single mouse. Arrows indicate the time of morpholino injection. (b) Individual data points for Fig. 4e, f were plotted. Each line represents Epo protein concentrations (red line) or hematocrit counts (blue line) measured at the indicted time points for a single mouse. Arrows indicate the time of morpholino injection.
Supplementary Figure 9 Intravenous injections of high doses of v-M8 morpholino do not affect mouse body weight.
(a) Nineteen-week old female BALB/c mice were i.v. injected with v-M8 morpholino at 0, 2.5, 5.0, or 10.0 mg/kg/day for two days (n = 5, 4, 5, and 5 mice, respectively). Mouse body weight was measured at the indicated time points. Arrows indicate v-M8 injections. Each line represents body weight data collected from a single mouse. No significant change on body weight was observed for mice treated with different doses of v-M8 morpholino (paired-sample Student’s t-test, two-sided, P = 0.07, 0.09, and 0.34, respectively). (b) Experiments similar to (a) except that eight-week old female BALB/c mice were i.v. injected with v-M8 morpholino at 0 or 12.5 mg/kg/day for four days (n = 10 and 10 mice, respectively). No significant change on body weight was observed for the two mouse groups (paired-sample Student’s t-test, two-sided, P = 0.59).
Supplementary information
Supplementary Information
Supplementary Figs. 1–9.
Rights and permissions
About this article
Cite this article
Zhong, G., Wang, H., He, W. et al. A reversible RNA on-switch that controls gene expression of AAV-delivered therapeutics in vivo. Nat Biotechnol 38, 169–175 (2020). https://doi.org/10.1038/s41587-019-0357-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0357-y
This article is cited by
-
Adeno-associated virus as a delivery vector for gene therapy of human diseases
Signal Transduction and Targeted Therapy (2024)
-
Engineered poly(A)-surrogates for translational regulation and therapeutic biocomputation in mammalian cells
Cell Research (2024)
-
Ribo-On and Ribo-Off tools using a self-cleaving ribozyme allow manipulation of endogenous gene expression in C. elegans
Communications Biology (2023)
-
Customizing cellular signal processing by synthetic multi-level regulatory circuits
Nature Communications (2023)
-
Prospective approaches to gene therapy computational modeling – spotlight on viral gene therapy
Journal of Pharmacokinetics and Pharmacodynamics (2023)