Abstract
Mitochondrial DNA (mtDNA) mutagenesis and nuclear DNA repair defects are considered cellular mechanisms of ageing. mtDNA mutator mice with increased mtDNA mutagenesis show signs of premature ageing. However, why patients with mitochondrial diseases, or mice with other forms of mitochondrial dysfunction, do not age prematurely remains unknown. Here, we show that cells from mutator mice display challenged nuclear genome maintenance similar to that observed in progeric cells with defects in nuclear DNA repair. Cells from mutator mice show slow nuclear DNA replication fork progression, cell cycle stalling and chronic DNA replication stress, leading to double-strand DNA breaks in proliferating progenitor or stem cells. The underlying mechanism involves increased mtDNA replication frequency, sequestering of nucleotides to mitochondria, depletion of total cellular nucleotide pools, decreased deoxynucleoside 5′-triphosphate (dNTP) availability for nuclear genome replication and compromised nuclear genome maintenance. Our data indicate that defects in mtDNA replication can challenge nuclear genome stability. We suggest that defects in nuclear genome maintenance, particularly in the stem cell compartment, represent a unified mechanism for mouse progerias. Therefore, through their destabilizing effects on the nuclear genome, mtDNA mutations are indirect contributors to organismal ageing, suggesting that the direct role of mtDNA mutations in driving ageing-like symptoms might need to be revisited.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
10 August 2020
A Correction to this paper has been published: https://doi.org/10.1038/s42255-020-0255-0
References
Harman, D. The biologic clock: the mitochondria? J. Am. Geriatr. Soc. 20, 145–147 (1972).
Trifunovic, A. et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423 (2004).
Kujoth, G. C. et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309, 481–484 (2005).
Trifunovic, A. et al. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc. Natl Acad. Sci. USA 102, 17993–17998 (2005).
Ahlqvist, K. J. et al. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell Metab. 15, 100–109 (2012).
Ahlqvist, K. J. et al. MtDNA mutagenesis impairs elimination of mitochondria during erythroid maturation leading to enhanced erythrocyte destruction. Nat. Commun. 6, 6494 (2015).
Hamalainen, R. H. et al. mtDNA mutagenesis disrupts pluripotent stem cell function by altering redox signaling. Cell Rep. 11, 1614–1624 (2015).
Norddahl, G. L. et al. Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. Cell Stem Cell 8, 499–510 (2011).
Carrero, D., Soria-Valles, C. & Lopez-Otin, C. Hallmarks of progeroid syndromes: lessons from mice and reprogrammed cells. Dis. Model Mech. 9, 719–735 (2016).
Gorman, G. S. et al. Mitochondrial diseases. Nat. Rev. Dis. Prim. 2, 16080 (2016).
Tyynismaa, H. et al. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc. Natl Acad. Sci. USA 102, 17687–17692 (2005).
Behrens, A., van Deursen, J. M., Rudolph, K. L. & Schumacher, B. Impact of genomic damage and ageing on stem cell function. Nat. Cell Biol. 16, 201–207 (2014).
Ito, K. et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature 431, 997–1002 (2004).
Iyer, D. R. & Rhind, N. Replication fork slowing and stalling are distinct, checkpoint-independent consequences of replicating damaged DNA. PLoS Genet. 13, e1006958 (2017).
Barlow, C. et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 86, 159–171 (1996).
Reichard, P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev. Biochem. 57, 349–374 (1988).
Meuth, M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp. Cell Res. 181, 305–316 (1989).
Nishigaki, Y., Marti, R., Copeland, W. C. & Hirano, M. Site-specific somatic mitochondrial DNA point mutations in patients with thymidine phosphorylase deficiency. J. Clin. Invest 111, 1913–1921 (2003).
Gandhi, V. V. & Samuels, D. C. A review comparing deoxyribonucleoside triphosphate (dNTP) concentrations in the mitochondrial and cytoplasmic compartments of normal and transformed cells. Nucleosides Nucleotides Nucleic Acids 30, 317–339 (2011).
Tubbs, A. et al. Dual roles of poly(dA:dT) tracts in replication initiation and fork collapse. Cell 174, 1127–1142 (2018).
Pai, C. C. & Kearsey, S. E. A critical balance: dNTPs and the maintenance of genome stability. Genes (Basel) 8, 57 (2017).
Rampazzo, C. et al. Regulation by degradation, a cellular defense against deoxyribonucleotide pool imbalances. Mutat. Res. 703, 2–10 (2010).
Blazquez-Bermejo, C. et al. Increased dNTP pools rescue mtDNA depletion in human POLG-deficient fibroblasts. FASEB J. 33, 7168–7179 (2019).
Nilsson, R. et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 5, 3128 (2014).
Macao, B. et al. The exonuclease activity of DNA polymerase gamma is required for ligation during mitochondrial DNA replication. Nat. Commun. 6, 7303 (2015).
Reyes, A. et al. Mitochondrial DNA replication proceeds via a ‘bootlace’ mechanism involving the incorporation of processed transcripts. Nucleic Acids Res. 41, 5837–5850 (2013).
Torregrosa-Munumer, R., Goffart, S., Haikonen, J. A. & Pohjoismaki, J. L. Low doses of ultraviolet radiation and oxidative damage induce dramatic accumulation of mitochondrial DNA replication intermediates, fork regression, and replication initiation shift. Mol. Biol. Cell 26, 4197–4208 (2015).
Kaufman, B. A. et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell 18, 3225–3236 (2007).
Ylikallio, E., Tyynismaa, H., Tsutsui, H., Ide, T. & Suomalainen, A. High mitochondrial DNA copy number has detrimental effects in mice. Hum. Mol. Genet. 19, 2695–2705 (2010).
Jiang, M. et al. Increased total mtDNA copy number cures male infertility despite unaltered mtDNA mutation load. Cell Metab. 26, 429–436 (2017).
Wheaton, K. et al. Progerin-induced replication stress facilitates premature senescence in Hutchinson-Gilford progeria syndrome. Mol. Cell Biol. 37, e00659–16 (2017).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Lopez, L. C. et al. Unbalanced deoxynucleotide pools cause mitochondrial DNA instability in thymidine phosphorylase-deficient mice. Hum. Mol. Genet. 18, 714–722 (2009).
Carpenter, A. E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Handel, M. A. The XY body: a specialized meiotic chromatin domain. Exp. Cell Res. 296, 57–63 (2004).
Marti, R., Dorado, B. & Hirano, M. Measurement of mitochondrial dNTP pools. Methods Mol. Biol. 837, 135–148 (2012).
Landoni, J. C., Wang, L. & Suomalainen, A. Quantitative solid-phase assay to measure deoxynucleoside triphosphate pools. Biol. Methods Protoc. 3, bpy011 (2018).
Palacino, J. J. et al. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J. Biol. Chem. 279, 18614–18622 (2004).
Icay, K. et al. SePIA: RNA and small RNA sequence processing, integration, and analysis. BioData Min. 9, 20 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Tarca, A. L. et al. A novel signaling pathway impact analysis. Bioinformatics 25, 75–82 (2009).
Acknowledgements
We thank T. Manninen, H. Ojala, M. Innilä and A. Muranen (University of Helsinki) for technical assistance, C. Storgaard Sørensen and K. Voßgröne (University of Copenhagen) for technical advice and T. McWilliams, C. Dunn and K. Wartiovaara (University of Helsinki) for discussion and critical comments. This work was supported by the Academy of Finland (275215 to R.H.H., 307592 to A.S., 303349 A.S., 30743 to A.S.), the Sigrid Jusélius Foundation, Jane and Aatos Erkko Foundation, the University of Helsinki, the University of Eastern Finland and the European Research Council (268955 to A.S.).
Author information
Authors and Affiliations
Contributions
R.H.H. was responsible for study conception and design, experimental work, data analysis and interpretation, and writing of the manuscript. J.C.L., K.J.A., S.R., M.O.R., S.G. and L.W. did the experimental work and data analysis. V.B., K.I. and S.H. did the bioinformatics and data analysis. M.L. was responsible for study design and data interpretation. A.S. was responsible for study conception and design, data interpretation and writing of manuscript. All authors commented on and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Primary Handling Editor: Christoph Schmitt.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
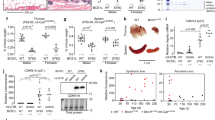
Extended Data Fig. 1 Increased DNA damage in mtDNA mutator mouse embryonic fibroblasts.
Mean fluorescence intensity (MFI) of γH2AX staining per cell; quantification of FACS signals and representative histograms. WT n = 4, Mut n = 4, three independent experiments, P = 0.0287. WT, black dots; mutator, red squares. Data represented as mean ± s.e.m. and analysed with unpaired t-test. n corresponds to biological replicates.
Extended Data Fig. 2 Increased DNA breaks in primary spermatocytes in mutator testes.
Fraction of γH2AX positive nuclei in primary spermatocytes; quantification of signals and representative original and processed images from γH2AX (red) and DAPI (blue) stainings. WT n = 5, mutator n = 7, approximately 1,000 nuclei per mouse, P = 0.0376. Scale bars, 100 μm. WT, black dots; Mut, red squares. Data represented as mean ± s.e.m. and analysed with unpaired t-test. n corresponds to biological replicates.
Extended Data Fig. 3 Decreased dNTP pools in mtDNA mutator mouse embryonic fibroblasts.
a, Total cellular nucleotide pools, WT n = 3, Mut n = 3, three independent measurements. b, Quantification of individual nucleotides. WT n = 3, Mut n = 3, three independent measurements. WT, black dots; Mut, red squares. Data presented as mean ± s.e.m. and analysed with unpaired t-test. n corresponds to biological replicates.
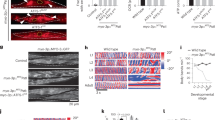
Extended Data Fig. 4 Supplementation with nucleosides does not increase proliferation nor significantly reduce DNA breaks in mutator iPSCs.
a, Growth curves of WT and mutator cells untreated (solid lines) and supplemented with nucleosides (C-73, G-85, U-73, A-80, T-24 mg/l) (dashed lines), n = 4, three independent experiments. b, Relative mean fluorescence intensity (MFI) of γH2AX staining per cell compared to the untreated cells; quantification of FACS signals. The 40 h treatments with excess nucleosides do not have any effect on WT cells. Slight reduction on γH2AX staining in mutator cells was only suggestive. WT n = 4, Mut n = 5, three independent experiments. WT, black dots; Mut, red squares. All data represented as mean ± s.e.m. and analysed with unpaired t-test. n corresponds to biological replicates.
Extended Data Fig. 5 Relative expression of 5′nucleotidase genes.
WT n = 3, Mut n = 5. Nt5e P = 0.0518, Nt5dc1 P = 0.0176, Nt5dc3 P = 0.00258. WT, black dots; Mut, red squares. All data represented as mean ± s.e.m. and analysed with unpaired t-test. n corresponds to biological replicates.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Table 1
Source data
Rights and permissions
About this article
Cite this article
Hämäläinen, R.H., Landoni, J.C., Ahlqvist, K.J. et al. Defects in mtDNA replication challenge nuclear genome stability through nucleotide depletion and provide a unifying mechanism for mouse progerias. Nat Metab 1, 958–965 (2019). https://doi.org/10.1038/s42255-019-0120-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-019-0120-1
This article is cited by
-
Mitochondrial stress: a key role of neuroinflammation in stroke
Journal of Neuroinflammation (2024)
-
Mitochondrial complex III deficiency drives c-MYC overexpression and illicit cell cycle entry leading to senescence and segmental progeria
Nature Communications (2023)
-
OxPhos defects cause hypermetabolism and reduce lifespan in cells and in patients with mitochondrial diseases
Communications Biology (2023)
-
Biomarkers of aging
Science China Life Sciences (2023)
-
Cellular pyrimidine imbalance triggers mitochondrial DNA–dependent innate immunity
Nature Metabolism (2021)