Abstract
Enteric pathogens are exposed to a dynamic polymicrobial environment in the gastrointestinal tract1. This microbial community has been shown to be important during infection, but there are few examples illustrating how microbial interactions can influence the virulence of invading pathogens2. Here we show that expansion of a group of antibiotic-resistant, opportunistic pathogens in the gut—the enterococci—enhances the fitness and pathogenesis of Clostridioides difficile. Through a parallel process of nutrient restriction and cross-feeding, enterococci shape the metabolic environment in the gut and reprogramme C. difficile metabolism. Enterococci provide fermentable amino acids, including leucine and ornithine, which increase C. difficile fitness in the antibiotic-perturbed gut. Parallel depletion of arginine by enterococci through arginine catabolism provides a metabolic cue for C. difficile that facilitates increased virulence. We find evidence of microbial interaction between these two pathogenic organisms in multiple mouse models of infection and patients infected with C. difficile. These findings provide mechanistic insights into the role of pathogenic microbiota in the susceptibility to and the severity of C. difficile infection.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Raw RNA-sequencing data and processed data have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE165751. MGE clusters identified in Extended Data Fig. 2c are available in Supplementary Table 4. Conservation of arc genes performed on metagenomic reads has been deposited under BioProject ID PRJNA74826228. Conservation of arc genes among enterococci performed on dataset has been deposited under BioProject ID PRJNA58716125,26. Source data are provided with this paper.
Code availability
Code and source data for replication of the C. difficile GENRE (iCdR703) analyses are available at https://github.com/mjenior/Smith_etal_Enterococcus.
References
Abbas, A. & Zackular, J. P. Microbe–microbe interactions during Clostridioides difficile infection. Curr. Opin. Microbiol. 53, 19–25 (2020).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015).
Lessa, F. C., Winston, L. G. & McDonald, L. C., Team, E. I. P. C. d. S. Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 372, 2369–2370 (2015).
Schubert, A. M. et al. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. mBio 5, e01021–01014 (2014).
Auchtung, J. M., Preisner, E. C., Collins, J., Lerma, A. I. & Britton, R. A. Identification of simplified microbial communities that inhibit Clostridioides difficile infection through dilution/extinction. mSphere 5, e00387–20 (2020).
Zackular, J. P. et al. Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nat. Med. 22, 1330–1334 (2016).
Tomkovich, S., Stough, J. M. A., Bishop, L. & Schloss, P. D. The initial gut microbiota and response to antibiotic perturbation influence Clostridioides difficile clearance in mice. mSphere 5, e00869–20 (2020).
Berkell, M. et al. Microbiota-based markers predictive of development of Clostridioides difficile infection. Nat. Commun. 12, 2241 (2021).
Antharam, V. C. et al. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 51, 2884–2892 (2013).
Poduval, R. D., Kamath, R. P., Corpuz, M., Norkus, E. P. & Pitchumoni, C. S. Clostridium difficile and vancomycin-resistant Enterococcus: the new nosocomial alliance. Am. J. Gastroenterol. 95, 3513–3515 (2000).
Ubeda, C. et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 120, 4332–4341 (2010).
Taur, Y. et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis. 55, 905–914 (2012).
Willett, J. L. E. et al. Comparative biofilm assays using Enterococcus faecalis OG1RF identify new determinants of biofilm formation. mBio 12, e0101121 (2021).
Willett, J. L. E., Robertson, E. B. & Dunny, G. M. The phosphatase Bph and peptidyl–prolyl isomerase PrsA are required for gelatinase expression and activity in Enterococcus faecalis. J. Bacteriol. 204, e0012922 (2022).
Lee, I. P. A., Eldakar, O. T., Gogarten, J. P. & Andam, C. P. Bacterial cooperation through horizontal gene transfer. Trends Ecol. Evol. 37, 223–232 (2021).
Roberts, A. P. & Mullany, P. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol. Rev. 35, 856–871 (2011).
Chambers, C. J., Roberts, A. K., Shone, C. C. & Acharya, K. R. Structure and function of a Clostridium difficile sortase enzyme. Sci. Rep. 5, 9449 (2015).
Jenior, M. L. et al. Novel drivers of virulence in Clostridioides difficile identified via context-specific metabolic network analysis. mSystems 6, e0091921 (2021).
Fang, X., Lloyd, C. J. & Palsson, B. O. Reconstructing organisms in silico: genome-scale models and their emerging applications. Nat. Rev. Microbiol. 18, 731–743 (2020).
Jenior, M. L., Moutinho, T. J., Dougherty, B. V. & Papin, J. A. Transcriptome-guided parsimonious flux analysis improves predictions with metabolic networks in complex environments. PLoS Comput. Biol. 16, e1007099 (2020).
Pruss, K. M. et al. Oxidative ornithine metabolism supports non-inflammatory C. difficile colonization. Nat Metab 4, 19–28 (2022).
Barker, H. A. Amino acid degradation by anaerobic bacteria. Annu. Rev. Biochem. 50, 23–40 (1981).
Matthews, M. L. et al. Chemoproteomic profiling and discovery of protein electrophiles in human cells. Nat. Chem. 9, 234–243 (2017).
Keogh, D. et al. Enterococcal metabolite cues facilitate interspecies niche modulation and polymicrobial infection. Cell Host Microbe 20, 493–503 (2016).
Sundermann, A. J. et al. Whole genome sequencing surveillance and machine learning of the electronic health record for enhanced healthcare outbreak detection. Clin. Infect. Dis. 75, 476–482 (2021).
Bryan, N. C. et al. Genomic and functional characterization of Enterococcus faecalis isolates recovered from the International Space Station and their potential for pathogenicity. Front. Microbiol. 11, 515319 (2020).
Deibel, R. H. Utilization of arginine as an energy source for the growth of Streptococcus faecalis. J. Bacteriol. 87, 988–992 (1964).
Fishbein, S. R. et al. Multi-omics investigation of Clostridioides difficile-colonized patients reveals pathogen and commensal correlates of C. difficile pathogenesis. eLife 11, e72801 (2022).
Karasawa, T., Maegawa, T., Nojiri, T., Yamakawa, K. & Nakamura, S. Effect of arginine on toxin production by Clostridium difficile in defined medium. Microbiol. Immunol. 41, 581–585 (1997).
Fredrick, C. M., Lin, G. & Johnson, E. A. Regulation of botulinum neurotoxin synthesis and toxin complex formation by arginine and glucose in Clostridium botulinum ATCC 3502. Appl. Environ. Microbiol. 83, e00642–17 (2017).
Bushman, F. D. et al. Multi-omic analysis of the interaction between Clostridioides difficile infection and pediatric inflammatory bowel disease. Cell Host Microbe 28, 422–433.e7 (2020).
Keith, J. W. et al. Impact of antibiotic-resistant bacteria on immune activation and Clostridioides difficile infection in the mouse intestine. Infect. Immun. 88, e00362–19 (2020).
Lesniak, N. A. et al. The gut bacterial community potentiates Clostridioides difficile infection severity. mBio 13, e0118322 (2022).
Girinathan, B. P. et al. In vivo commensal control of Clostridioides difficile virulence. Cell Host Microbe 29, 1693–1708.e1697 (2021).
Hirose, Y. et al. Streptococcus pyogenes upregulates arginine catabolism to exert its pathogenesis on the skin surface. Cell Rep. 34, 108924 (2021).
Stabler, R. A. et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10, R102 (2009).
Carter, G. P. et al. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections. mBio 6, e00551 (2015).
Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 (1983).
Dale, J. L. et al. Comprehensive functional analysis of the Enterococcus faecalis core genome using an ordered, sequence-defined collection of insertional mutations in strain OG1RF. mSystems 3, e00062–18 (2018).
Theriot, C. M. et al. Cefoperazone-treated mice as an experimental platform to assess differential virulence of Clostridium difficile strains. Gut Microbes 2, 326–334 (2011).
Kumar, L., Cox, C. R. & Sarkar, S. K. Matrix metalloprotease-1 inhibits and disrupts Enterococcus faecalis biofilms. PLoS One 14, e0210218 (2019).
Bloedt, K., Riecker, M., Poppert, S. & Wellinghausen, N. Evaluation of new selective culture media and a rapid fluorescence in situ hybridization assay for identification of Clostridium difficile from stool samples. J. Med. Microbiol. 58, 874–877 (2009).
Wellinghausen, N., Bartel, M., Essig, A. & Poppert, S. Rapid identification of clinically relevant Enterococcus species by fluorescence in situ hybridization. J. Clin. Microbiol. 45, 3424–3426 (2007).
Knippel, R. J. et al. Heme sensing and detoxification by HatRT contributes to pathogenesis during Clostridium difficile infection. PLoS Pathog. 14, e1007486 (2018).
Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003).
Calle, M. L., Urrea, V., Boulesteix, A. L. & Malats, N. AUC-RF: a new strategy for genomic profiling with random forest. Hum. Hered. 72, 121–132 (2011).
Hankin, J. A., Barkley, R. M. & Murphy, R. C. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass. Spectrom. 18, 1646–1652 (2007).
Thomas, A., Charbonneau, J. L., Fournaise, E. & Chaurand, P. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition. Anal. Chem. 84, 2048–2054 (2012).
Yang, J. & Caprioli, R. M. Matrix sublimation/recrystallization for imaging proteins by mass spectrometry at high spatial resolution. Anal. Chem. 83, 5728–5734 (2011).
Prentice, B. M. et al. Dynamic range expansion by gas-phase ion fractionation and enrichment for imaging mass spectrometry. Anal. Chem. 92, 13092–13100 (2020).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Jackson, S., Calos, M., Myers, A. & Self, W. T. Analysis of proline reduction in the nosocomial pathogen Clostridium difficile. J. Bacteriol. 188, 8487–8495 (2006).
Evans, D. R. et al. Systematic detection of horizontal gene transfer across genera among multidrug-resistant bacteria in a single hospital. eLife 9, e53886 (2020).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Bryan, E. M., Bae, T., Kleerebezem, M. & Dunny, G. M. Improved vectors for nisin-controlled expression in Gram-positive bacteria. Plasmid 44, 183–190 (2000).
Chilambi, G. S. et al. Evolution of vancomycin-resistant Enterococcus faecium during colonization and infection in immunocompromised pediatric patients. Proc. Natl Acad. Sci. USA 117, 11703–11714 (2020).
Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Obeid, J. S. et al. Procurement of shared data instruments for Research Electronic Data Capture (REDCap). J. Biomed. Inform. 46, 259–265 (2013).
Acknowledgements
We thank L. Harrison and J. Marsh for sharing genome sequence data and VRE isolates from the EDS-HAT study (R01AI127472); the Children’s Hospital of Philadelphia Junior Faculty Breakfast Club and the Children’s Hospital of Philadelphia MICRO group for feedback on the manuscript; all members of the Zackular laboratory for providing feedback and support; the Children’s Hospital of Philadelphia Biostatistics and Data Management Core for statistical consulting and support; E. Friedman and the Penn Microbial Culture and Metabolomics Core for their support and expertise; and K. Bittinger and C. Tanes for thoughtful discussions on analyses detailed in this study. Funding was provided by the National Institutes of Health grant K22AI7220 (J.P.Z.), National Institutes of Health grant R35GM138369 (J.P.Z.), Children’s Hospital of Philadelphia Junior Faculty Pilot Grant (J.P.Z.), Cell and Molecular Biology Training Grant T32GM07229 (A.B.S.), National Institutes of Health grant R01AI138581 (E.P.S.), National Institutes of Health grant R01AI145992 (E.P.S.), National Institutes of Health grant R01AI164587 (E.P.S.), UVA TransUniversity Microbiome Initiative Pilot Grant (M.L.J. and J.A.P.), National Institutes of Health grant R01AT010253 (J.A.P.), Chemical and Biology Interface Training Grant 5T32GM071339-15 (K.A.B.), National Institutes of Health grant R01HD090061 (J.A.G.), National Institutes of Health/National Center for Advancing Translational Sciences Grant Support (UL1TR000445) for REDCap (Vanderbilt University) (M.R.N.), National Institutes of Health grant K23AI156132-01 (M.R.N.), National Institutes of Health grant K23 AI121485 (B.J.K.), Centers for Disease Control and Prevention grant BAA 200-2016-91937 (B.J.K.), Perelman School of Medicine/Children’s Hospital of Philadelphia PennCHOP Microbiome Program (R.N.B.), Crohn’s and Colitis Foundation (R.N.B.), Penn Center for AIDS Research P30-AI045008 (R.N.B.) and Commonwealth Universal Research Enhancement programme grant SAP#4100068710 (R.N.B.)
Author information
Authors and Affiliations
Contributions
A.B.S., E.P.S. and J.P.Z. conceived the study. A.B.S. and J.P.Z designed and performed experimentation and mouse work with support from O.K. P.C.R., J.L.H., C.L., J.G., M.A.S. and J.A.G. B.M.P. and J.S. performed the imaging mass spectrometry and analyses. K.A.B. and M.L.M. developed the hydrazine probe and performed corresponding analyses. B.J.K., F.D.B., J.E., R.N.B. and M.R.N. provided human samples and related analyses. M.L.J. and J.A.P. developed C. difficile genome-scale metabolic network reconstructions. A.A. and C.D. supported data analyses and bioinformatics. E.E.F. performed pathology analyses. D.R.E. and D.V.T. performed the MGE analyses and collagen-binding assay. Transposon library and biofilm support was provided by G.M.D. Funding was acquired by E.P.S., B.M.P., M.L.J., M.L.M., J.A.P., J.A.G., M.R.N., B.J.K., A.B.S. and J.P.Z. The Article was written by A.B.S. and J.P.Z. with input from the other authors. J.P.Z supervised the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Robert Britton and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Enterococcal abundance and dynamics during CDI.
(a) Enterococcal bacterial burdens (CFUs) from pediatric patients (black and gold = Vanderbilt University (median, n = 24 for Non-CDI controls; n = 34 for CDI, two-sided Mann-Whitney, P = 0.004); blue and light blue = Children’s Hospital of Philadelphia (n = 19 for healthy; n = 20 for IBD + CDI, two-sided Mann-Whitney test, P = 0.012). (b) Two-sided Spearman correlation between detected C. difficile and Enterococcus burdens in pediatric patients with IBD + CDI (Spearman ρ = 0.551; n = 19). (c) Bacterial burdens quantified from mice following treatment with cefoperazone (cef) or cef + vancomycin (vanc) (mean ± s.e.m., n = 10 mice/group). (d) Enterococcal CFUs over the course of CDI. Mice were infected with a toxin producing wild type strain (M7404 TcdA+TcdB+) or a toxin-null isogenic mutant (M7404 TcdA-TcdB-) (n = 5/group) (mean ± s.e.m. two-sided Mann-Whitney with Bonferroni-Dunn method for correction for multiple comparisons, corrected P values are in Supplementary Table 5).
Extended Data Fig. 2 Biofilm formation and transfer of mobile genetic elements during interspecies interactions.
(a) Survival assay of co-culture biofilms with E. faecalis (E.f.) (P = 0.119) or transposon mutants in E. faecalis genes OG1RF_11528 (fsrB::Tn) (P < 0.001) and OG1RF_10423 (prsA::Tn) (P < 0.001). Abundance of C. difficile in untreated (-) or vancomycin treated (+) biofilms are depicted (mean ± s.d., n = 7, two-sided Mann-Whitney with Bonferroni-Dunn method for correction for multiple comparisons) (b) Abundance of C. difficile (C.d.) and E. faecalis strains (E.f., prsA::Tn, and frsB::Tn) in untreated dual species biofilms (mean ± s.d., n = 3). (c) Clusters of shared sequences detected in C. difficile (blue) and VRE (olive) genomes of clinical isolates from hospitalized patients. Lines connect sequences with at least 99.98% identity. Clusters are labeled based on mobile element type and relevant cargo, if known. Source data for each cluster can be found in Supplementary Information. (d) Biofilm formation of E. faecalis OG1RF carrying empty pMSP3535 vector or pMSP3535 carrying the CD0386-like adhesin. Biofilm formation was tested in standard (P = 0.302) and collagen-coated plates (P<0.001). Crystal violet staining (OD550) values were calculated (mean ± s.d., n = 24/group for standard plates and 16/group for collagen plates, unpaired two-tailed t-tests).
Extended Data Fig. 3 Enterococcal-mediated enhancement of C. difficile toxin gene expression and production.
(a) Fold change of the toxin-encoding genes in C. difficile in coculture with E. faecalis versus monoculture as measured by qPCR (mean ± s.d., n = 3). (b) Toxin production by C. difficile when grown in co-culture with E. faecalis as measured by ELISA. Both C. difficile and E. faecalis were grown in the same culture and differential plating was used to measure C. difficile CFUs. Toxin levels (OD450) were normalized to C. difficile CFUs in the culture to control for any difference in growth (mean ± s.d., n = 5, two-tailed t-tests with Welch’s correction, P = 0.007). (c) C. difficile toxin levels measured from in vitro cultures by cytotoxicity with E. faecalis cell-free supernatants. (mean ± s.d., n = 5, Kruskal-Wallis test with Dunn’s correction for multiple comparisons, OG1RF P = 0.014, V583 P = 0.032). (d) C. difficile toxin production measured by cytotoxicity following introduction of cell-free supernatants from microbiota isolates cultured from human patients with CDI and IBD (mean ± s.d., n = 12 (C. difficile), 3 (Raoultella, Bifidobacterium, Enterobacter, Paeniclostridium, Lactobacillus), 5 (Klebsiella), 6 (Citrobacter, Clostridium, Shigella, Streptococcus), Kruskal-Wallis test with Dunn’s correction for multiple comparisons, Lactobacillus P = 0.049). Isolates were selected to represent the spectrum of taxa cultured from these patients.
Extended Data Fig. 4 Transcriptional changes associated with C. difficile – E. faecalis interactions.
(a) Pathway analysis of C. difficile transcripts significantly altered following co-culture as measured by RNA sequencing. For pathway analyses, blue bars represent transcripts that increased in abundance and red bars represent transcripts that decreased in abundance. (b) Volcano plot showing E. faecalis transcripts significantly altered following co-culture as measured by RNA sequencing. Red points represent genes associated with amino acid metabolism. Significance determined using two-sided Wald test and corrected for multiple comparisons using the Benjamini-Hochberg method. (c) Pathway analysis of E. faecalis transcripts significantly altered following co-culture as measured by RNA sequencing.
Extended Data Fig. 5 Transcriptome-guided metabolic flux predictions using genome-scale metabolic network reconstruction for C. difficile.
(a) AUC-Random Forest supervised machine learning results for reaction flux samples for conserved transport reactions between contexts (k = 10; OOB = 0%). (b) AUC-Random Forest supervised machine learning results for reaction flux samples for conserved amino acid transport reactions between contexts (k = 10; OOB = 0%). (c) Difference in simulated uptake of selected amino acids across context-specific models. Significance determined by two-sided Wilcoxon rank-sum test. Corrected P values in Supplementary Table 5.
Extended Data Fig. 6 In situ labelling of C. difficile with a hydrazine probe and gel-based profiling of D-proline reductase activity.
(a) Labeling schematic of hydrazine probe with PrdA of D-proline reductase. (b) Representative gel-based labelling profiles for C. difficile in the absence and presence of E. faecalis supernatant. Gel representative of three separate experiments. For Gel source data, see Supplementary Fig. 1. (c) Corresponding expression profiles after Coomassie staining.
Extended Data Fig. 7 The enterococcal ADI pathway reshapes the metabolic environment in the gut during CDI.
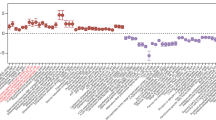
(a) C. difficile growth in presence of E. faecalis or E. faecalis arcD::Tn supernatants. (mean ± s.d., n = 8/group, two-way ANOVA, Time factor P < 0.001). (b) Taxonomic distribution (at bacterial family level) of reads mapped to arc genes in adult patients (n = 48) with symptomatic CDI. Each column is a subject and each row is a bacterial family. Each cell displays the percentage of reads mapped to arc genes of a specific family out of all arc mapped reads. (c) Relative abundance of the top 10 arc operon containing bacterial families in each adult patient symptomatically infected with C. difficile (n = 48, lower and upper hinges correspond to the first (25%) and third (75%) quartiles. The upper and lower whiskers extend from the hinge to the largest value no further than 1.5*IQR. Data beyond the whiskers are plotted individually). (d) Toxin production of C. difficile following introduction of supernatants from E. faecalis and addition of exogenous L-ornithine measured by ELISA (mean ± s.d., n = 3, Tukey’s multiple comparisons test, C. difficile +/- ornithine P = 0.705). (e) MALDI-IMS image of uninfected or infected mice (3d post-infection) (SPF) (representative of n = 5 mice) or (f) GF mice mono-infected with C. difficile CD196 or co-infected with E. faecalis OG1RF (2d post-infection) (representative of n = 4 mice). Individual heatmaps of arginine and ornithine. (g) Ornithine levels in stool measured by targeted metabolomics in GF mice infected with C. difficile only (mean ± s.d., n = 10) or C. difficile + E. faecalis OG1RF (mean ± s.d., n = 3, two-sided t-tests with Welch’s correction, P < 0.001). Metabolomics were performed on GF mice prior to infection (GF group, n = 13). (h) Arginine levels in stool of GF mice infected with C. difficile only (mean ± s.d., n = 10, two-sided t-test with Welch’s correction, P = 0.023) or C. difficile + E. faecalis OG1RF (mean ± s.d., n = 3, two-sided t-test with Welch’s correction, P<0.001). Stool metabolomics performed prior to infection (GF group, n = 13). (i) CFU of E. faecalis OG1RF (wild type) or E. faecalis arcD::Tn during CDI. Each strain introduced prior to CDI and naturally competed with endogenous enterococci (n = 5/group) (mean ± s.e.m. two-sided Mann-Whitney test with Bonferroni-Dunn method for correction for multiple comparisons, day 2 P = 0.048, day 3 P = 0.024). (j) Ornithine (P = 0.014) and (k) arginine (P<0.001) levels in stool measured by targeted metabolomics in GF mice infected with C. difficile. Mice pre-colonized for 1 day with E. faecalis (n = 3) or E. faecalis arcD::Tn (n = 4) (mean ± s.d., two-sided t-tests with Welch’s correction). Metabolomics performed on GF mice prior to infection (GF group, n = 5).
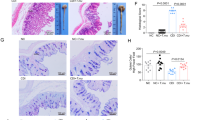
Extended Data Fig. 8 Arginine supplementation decreases C. difficile pathogenesis in mice.
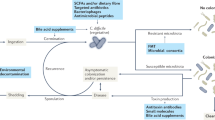
(a) C. difficile and (b) Enterococcus burdens quantified from mice following cefoperazone treatment and subsequent infection. Mice were treated with 2% L-arginine in drinking water starting 2 days prior to infection and subsequently during the course of infection (mean ± s.d., n = 7 for control, n = 8 for L-arginine treated; Mann-Whitney with Bonferroni-Dunn method for correction for multiple comparisons for each comparison). (c) Inflammation score (P = 0.023) and (d) cumulative pathology score (P = 0.051) measured 3 days post-infection for control (n = 7) and L-arginine treated (n = 8) mice (mean ± s.d., two-sided t-tests with Welch’s correction). (e) Spearman correlation between ornithine abundance in stool and C. difficile burdens in pediatric patients with IBD and CDI with detectable C. difficile based on culture (two-sided Spearman’s ρ = 0.4243; n = 26). (f) Proposed model of multifaceted cooperative interactions between enterococci and C. difficile during infection.
Supplementary information
Supplementary Information
This file contains Supplementary Fig. 1 and Tables 1–6.
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Smith, A.B., Jenior, M.L., Keenan, O. et al. Enterococci enhance Clostridioides difficile pathogenesis. Nature 611, 780–786 (2022). https://doi.org/10.1038/s41586-022-05438-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-05438-x
This article is cited by
-
The association between plasma free amino acids and type 2 diabetes mellitus complicated with infection in Chinese patients
Diabetology & Metabolic Syndrome (2024)
-
Promiscuous, persistent and problematic: insights into current enterococcal genomics to guide therapeutic strategy
BMC Microbiology (2024)
-
Gut diversity and the resistome as biomarkers of febrile neutropenia outcome in paediatric oncology patients undergoing hematopoietic stem cell transplantation
Scientific Reports (2024)
-
A commensal protozoan attenuates Clostridioides difficile pathogenesis in mice via arginine-ornithine metabolism and host intestinal immune response
Nature Communications (2024)
-
Vancomycin resistant enterococcus risk factors for hospital colonization in hematological patients: a matched case-control study
Antimicrobial Resistance & Infection Control (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.